Projects¶
EpiGraphDB¶
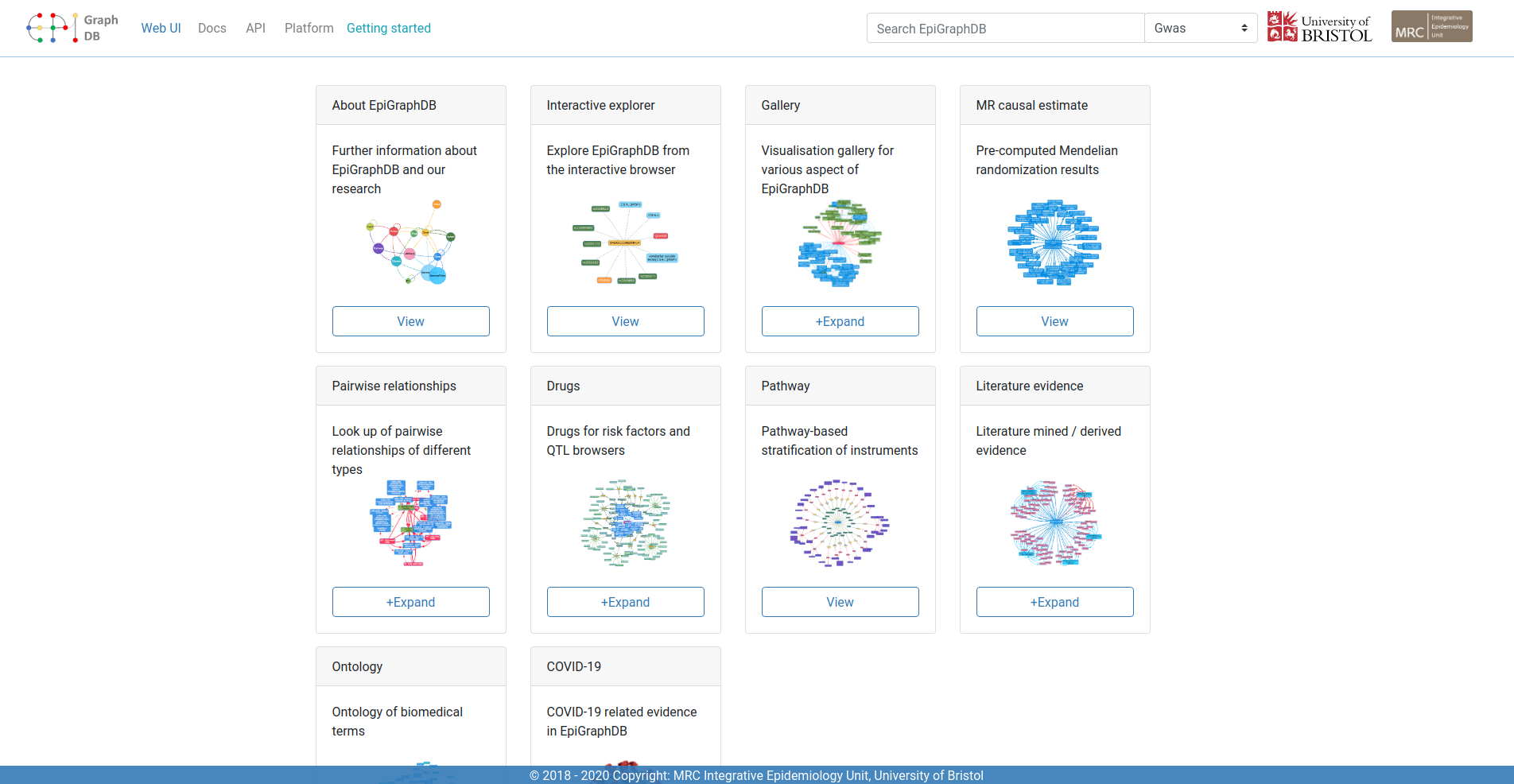
EpiGraphDB is an integrated knowledge graph on epidemiological evidence. Data in systematic statistical associations, literature, biological pathways, etc are integrated into a Neo4j graph database, which in combination with associated databases and other components support data mining of epidemiological relationships.
Read our paper on the EpiGraphDB platform here:
Liu, Y., Elsworth, B., Erola, P., Haberland, V., Hemani, G., Lyon, M., Zheng, J., Lloyd, O., Vabistsevits, M., Gaunt, T.R., 2020. EpiGraphDB: a database and data mining platform for health data science. Bioinformatics 37, 1304–1311. https://doi.org/10.1093/bioinformatics/btaa961
Read associated studies here:
Liu, Y., Gaunt, T.R., 2022. Triangulating evidence in health sciences with Annotated Semantic Queries. medRxiv. https://doi.org/10.1101/2022.04.12.22273803
Zheng, J., Zhang, Y., Zhao, H., Liu, Y., Baird, D., Karim, M.A., Ghoussaini, M., Schwartzentruber, J., Dunham, I., Elsworth, B., Roberts, K., Compton, H., Miller-Molloy, F., Liu, X., Wang, L., Zhang, H., Davey Smith, G., Gaunt, T.R., 2022. Multi-ancestry Mendelian randomization of omics traits revealing drug targets of COVID-19 severity. eBioMedicine 81, 104112. https://doi.org/10.1016/j.ebiom.2022.104112
Zhao, H., Rasheed, H., Nøst, T.H., Cho, Y., Liu, Y., Bhatta, L., Bhattacharya, A., Hemani, G., Davey Smith, G., Brumpton, B.M., Zhou, W., Neale, B.M., Gaunt, T.R., Zheng, J., 2022. Proteome-wide Mendelian randomization in global biobank meta-analysis reveals multi-ancestry drug targets for common diseases. Cell Genomics 2, 100195. https://doi.org/10.1016/j.xgen.2022.100195
Zheng, J., Haberland, V., Baird, D., Walker, V., Haycock, P.C., Hurle, M.R., Gutteridge, A., Erola, P., Liu, Y., Luo, S., Robinson, J., Richardson, T.G., Staley, J.R., Elsworth, B., Burgess, S., Sun, B.B., Danesh, J., Runz, H., Maranville, J.C., Martin, H.M., Yarmolinsky, J., Laurin, C., Holmes, M.V., Liu, J.Z., Estrada, K., Santos, R., McCarthy, L., Waterworth, D., Nelson, M.R., Smith, G.D., Butterworth, A.S., Hemani, G., Scott, R.A., Gaunt, T.R., 2020. Phenome-wide Mendelian randomization mapping the influence of the plasma proteome on complex diseases. Nat Genet. https://doi.org/10.1038/s41588-020-0682-6
Research in natural language processing¶
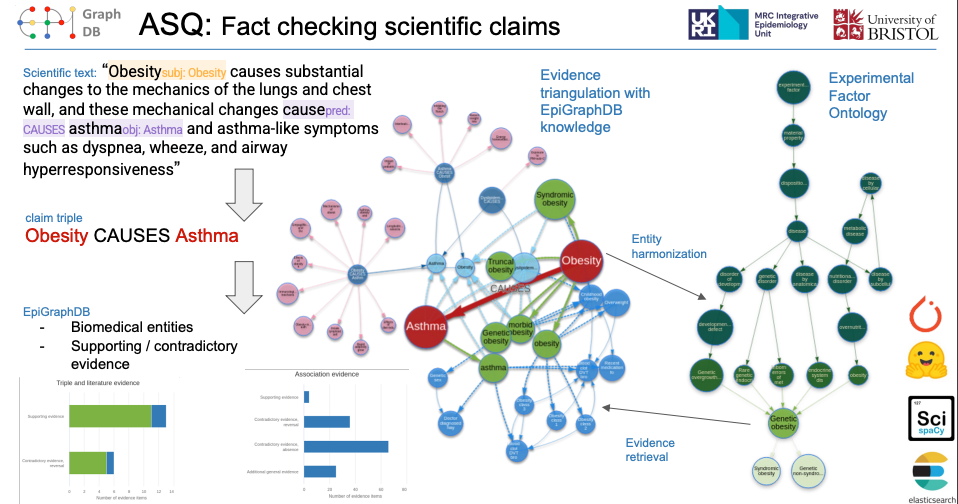
Originally within the context of EpiGraphDB, we have carried out a series of research studies using NLP to facilitate data mining.
Liu, Y., Gaunt, T.R., 2022. Triangulating evidence in health sciences with Annotated Semantic Queries. medRxiv. https://doi.org/10.1101/2022.04.12.22273803
Liu, Y., Elsworth, B.L., Gaunt, T.R., 2023. Using language models and ontology topology to perform semantic mapping of traits between biomedical datasets. Bioinformatics. https://doi.org/10.1093/bioinformatics/btad169
OpenGWAS¶
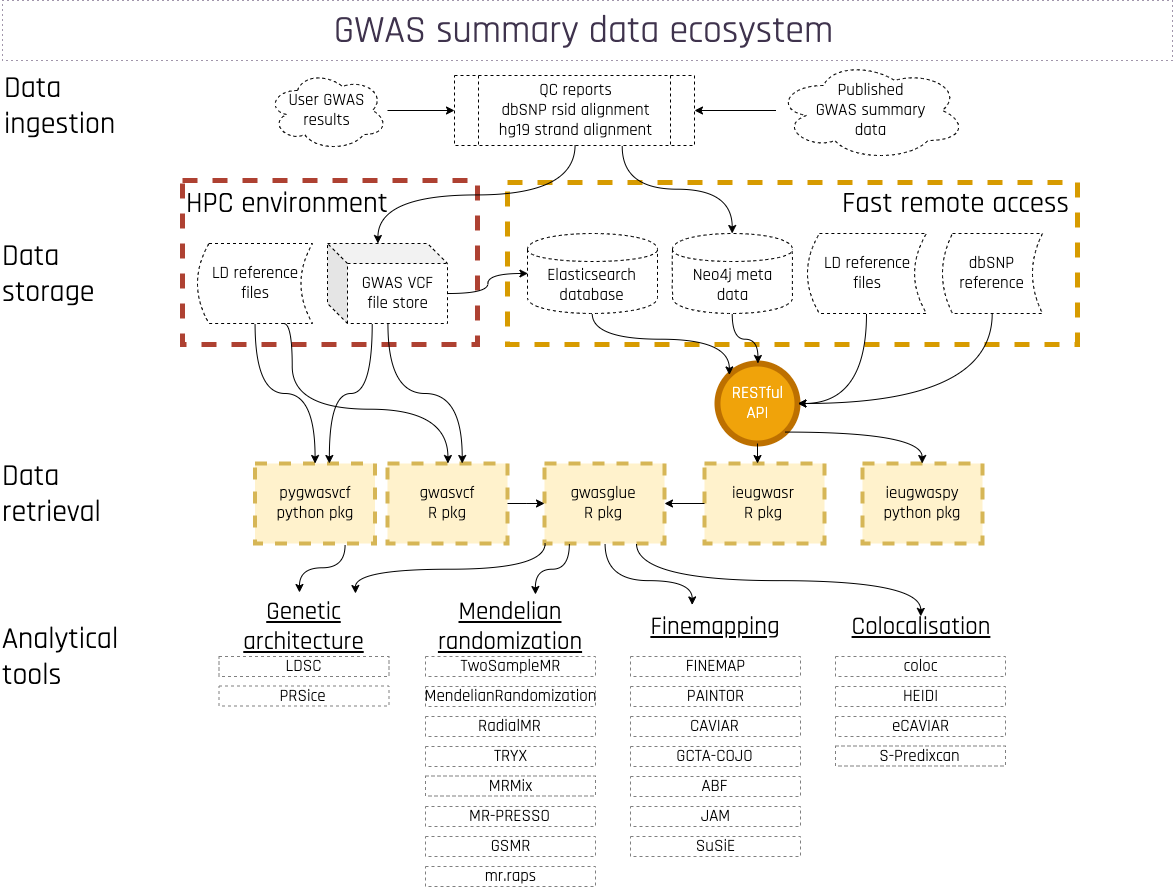
OpenGWAS is the data platform for currated collection of complete GWAS summary datasets, developed by MRC IEU memebers.
Elsworth, B., Lyon, M., Alexander, T., Liu, Y., Matthews, P., Hallett, J., Bates, P., Palmer, T., Haberland, V., Davey Smith, G., Zheng, J., Haycock, P., Gaunt, T.R., Hemani, G., 2020. The MRC IEU OpenGWAS data infrastructure. bioRxiv. https://doi.org/10.1101/2020.08.10.244293