Outline
- DeepWalk: intro to graph embeddings
- DeepWalk example
- Node2Vec
- DeepGL
- References
Graph and graph embeddings¶
$$
G = (V, E, X, Y)
$$$$
f(X) \rightarrow Y
$$
- $V$: vertices
- $E$: edges
- $X$: attributes
- $Y$: labels
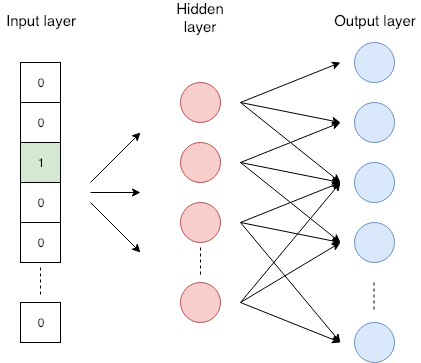
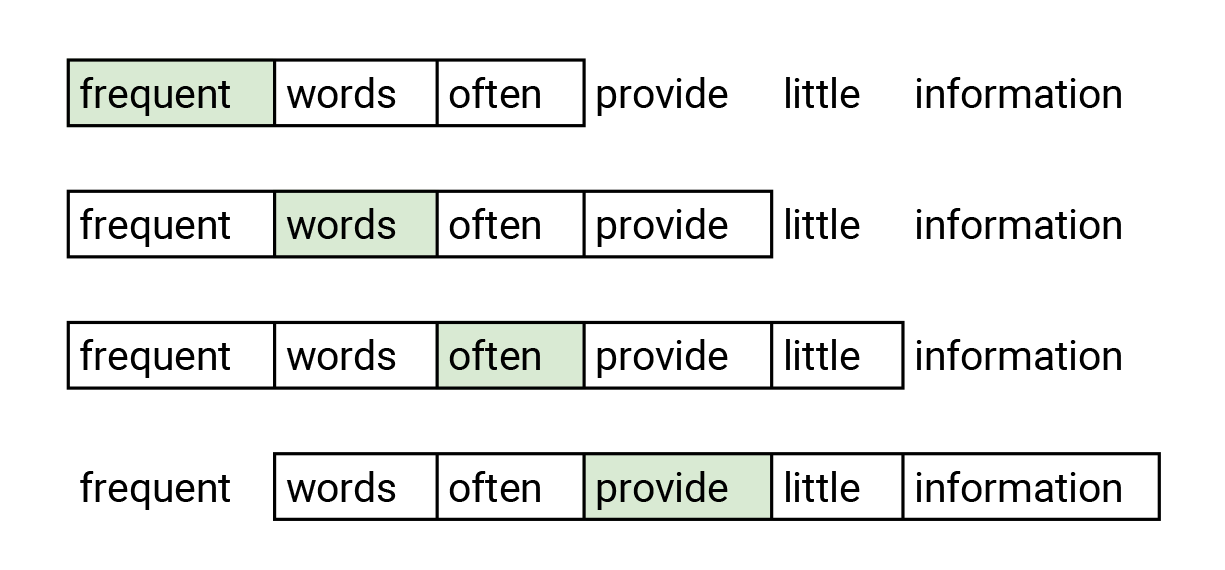
Word2Vec (SkipGram)¶
DeepWalk¶
From sentence embeddings to graph embeddings
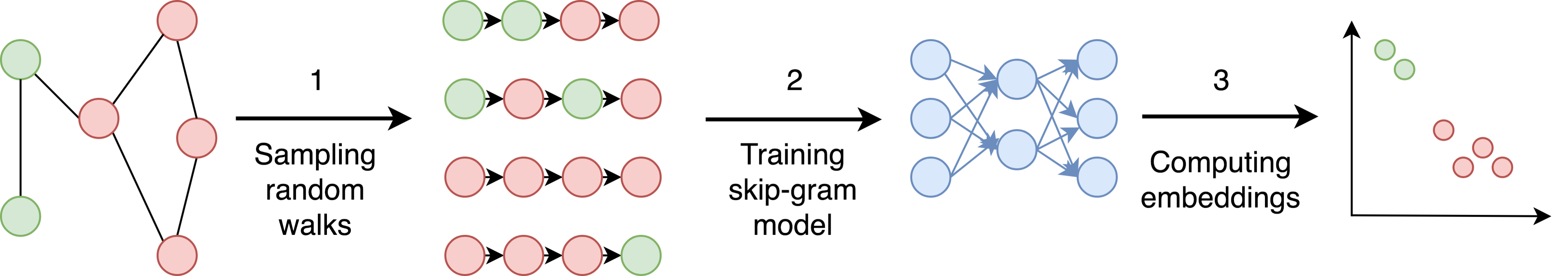
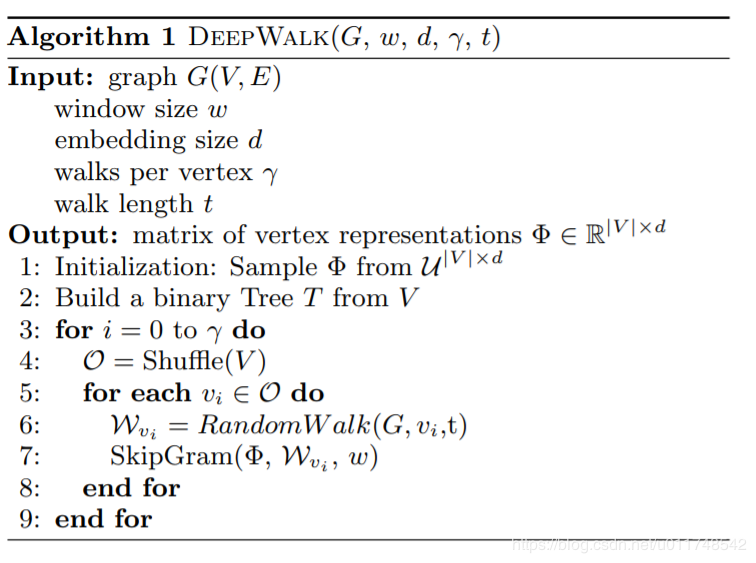
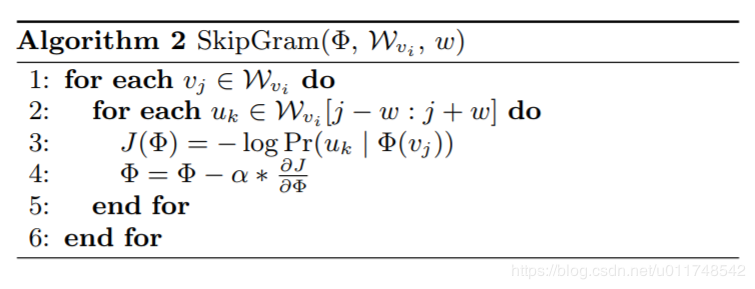

In [2]:
import numpy as np
import networkx as nx
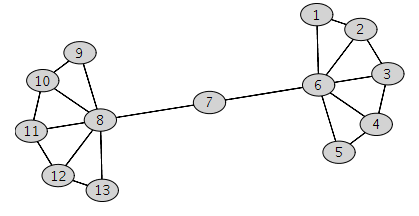
nx_graph = nx.Graph()
nodes = list(np.arange(1, 14))
# fmt: off
edges = [
(1, 2), (2, 3), (3, 4), (4, 5),
(1, 6), (2, 6), (3, 6), (4, 6), (5, 6),
(6, 7),
(7, 8),
(9, 8), (10, 8), (11, 8), (12, 8), (13, 8),
(9, 10), (10, 11), (11, 12), (12, 13),
]
# fmt: on
nx_graph.add_nodes_from(nodes)
nx_graph.add_edges_from(edges)
nx.draw(nx_graph, with_labels=True, node_color="white")
In [3]:
# https://github.com/phanein/deepwalk/blob/master/deepwalk/__main__.py
import random
import deepwalk
from deepwalk import walks as serialized_walks
from deepwalk.skipgram import Skipgram
from gensim.models import Word2Vec
from six import iterkeys
In [4]:
deepwalk_graph = deepwalk.graph.Graph()
for idx, x in enumerate(nx_graph.nodes()):
for y in iterkeys(nx_graph[x]):
deepwalk_graph[x].append(y)
deepwalk_graph.make_undirected()
deepwalk_graph
Out[4]:
Graph(list,
{1: [2, 6],
2: [1, 3, 6],
3: [2, 4, 6],
4: [3, 5, 6],
5: [4, 6],
6: [1, 2, 3, 4, 5, 7],
7: [6, 8],
8: [7, 9, 10, 11, 12, 13],
9: [8, 10],
10: [8, 9, 11],
11: [8, 10, 12],
12: [8, 11, 13],
13: [8, 12]})
In [5]:
# hyper-params
# num random walks per node: 80
NUM_WALKS = 80
# length of one random walk: 40
WALK_LENGTH = 40
# window size 10
WINDOW_SIZE = 10
# embedding dim 128
EMBEDDING_DIM = 128
In [6]:
walks = deepwalk.graph.build_deepwalk_corpus(
deepwalk_graph,
num_paths=NUM_WALKS,
path_length=WALK_LENGTH,
alpha=0,
rand=random.Random(123)
)
print(np.array(walks))
print(np.array(walks).shape)
[['9' '10' '11' ... '12' '13' '8'] ['6' '5' '6' ... '8' '10' '11'] ['8' '10' '11' ... '9' '10' '9'] ... ['13' '8' '10' ... '6' '4' '6'] ['6' '4' '5' ... '7' '8' '12'] ['8' '12' '13' ... '9' '10' '8']] (1040, 40)
In [7]:
model = Word2Vec(
walks,
size=EMBEDDING_DIM,
window=WINDOW_SIZE,
min_count=0,
sg=1,
hs=1,
workers=1,
)
In [8]:
# nodes, or "vocabulary"
model.wv.vocab
Out[8]:
{'9': <gensim.models.keyedvectors.Vocab at 0x1a1d7b6810>,
'10': <gensim.models.keyedvectors.Vocab at 0x1a1d7b6890>,
'11': <gensim.models.keyedvectors.Vocab at 0x1a1d7b68d0>,
'8': <gensim.models.keyedvectors.Vocab at 0x1a1d7b6910>,
'7': <gensim.models.keyedvectors.Vocab at 0x1a1d7b6990>,
'6': <gensim.models.keyedvectors.Vocab at 0x1a1d7b6a10>,
'1': <gensim.models.keyedvectors.Vocab at 0x1a1d7b6a50>,
'3': <gensim.models.keyedvectors.Vocab at 0x1a1d7b6a90>,
'2': <gensim.models.keyedvectors.Vocab at 0x1a1d7b6950>,
'4': <gensim.models.keyedvectors.Vocab at 0x1a1d7b69d0>,
'5': <gensim.models.keyedvectors.Vocab at 0x1a1d7b6ad0>,
'13': <gensim.models.keyedvectors.Vocab at 0x1a1d7b6b10>,
'12': <gensim.models.keyedvectors.Vocab at 0x1a1d7b6b50>}
In [9]:
# "word" embeddings
model.wv["1"]
Out[9]:
array([-0.00095934, -0.14086728, 0.07352262, 0.13970056, -0.0497438 ,
-0.2926532 , -0.06589775, -0.10685308, 0.18745854, -0.1012044 ,
-0.03285081, 0.29395282, -0.01746033, -0.04551164, -0.07437321,
0.00040195, 0.08093731, 0.04620257, 0.05116988, 0.02569078,
-0.12490681, 0.02504689, 0.1027611 , -0.08071101, -0.24925862,
0.12331013, -0.13090487, -0.1167615 , 0.15161267, 0.00957596,
-0.12357164, 0.08300231, 0.0844809 , -0.12852252, -0.20644854,
0.11501899, 0.26970205, 0.07551242, -0.11944196, -0.06687455,
-0.1480233 , -0.18081237, 0.17705128, -0.07423408, -0.08211829,
0.26821792, 0.0630434 , -0.23834868, -0.00506505, 0.07663959,
-0.08388641, 0.1582118 , -0.07193353, 0.01382565, 0.00668014,
0.01537236, -0.07911585, 0.09093268, 0.04782807, -0.04677264,
-0.23164956, -0.17346524, 0.12032165, -0.0476453 , -0.13943444,
-0.1260258 , 0.13450332, 0.10808325, -0.01826159, -0.3575986 ,
0.15718068, -0.04420409, -0.11785912, 0.10133453, -0.19064085,
-0.20066042, 0.08204125, 0.06170731, 0.16305035, -0.13007683,
-0.03484008, -0.14941491, -0.09808226, 0.08392007, 0.046721 ,
0.01600912, -0.07195069, 0.01543452, 0.13940337, -0.07913083,
-0.184816 , -0.0933992 , -0.07203937, -0.07271566, 0.11571857,
0.00552847, 0.00289924, -0.0523177 , 0.12037521, -0.09778723,
-0.03153525, -0.05874648, 0.08392701, 0.19099884, -0.00918258,
0.05699351, -0.04814161, -0.03177148, 0.00090648, -0.06779662,
-0.19197589, 0.12392153, 0.03059392, -0.21022129, -0.02727187,
0.13549924, 0.05740344, 0.00623336, -0.09875766, 0.04643035,
-0.10097225, 0.10052127, 0.06828507, 0.10290339, 0.1309135 ,
0.01477405, 0.08474117, 0.21653458], dtype=float32)
In [10]:
model.most_similar("1")
Out[10]:
[('3', 0.9698188304901123),
('6', 0.9323770999908447),
('2', 0.9054238796234131),
('4', 0.8182910084724426),
('5', 0.77535080909729),
('7', 0.6318682432174683),
('9', 0.05110911279916763),
('11', 0.05021246522665024),
('10', 0.03185462951660156),
('8', 0.02739347517490387)]
In [11]:
nx.draw(nx_graph, with_labels=True, node_color="white")
In [12]:
# Visualisation of "word" embeddings using PCA
from sklearn.decomposition import PCA
from matplotlib import pyplot
def plot_pca(model):
X = model[model.wv.vocab]
pca = PCA(n_components=2)
result = pca.fit_transform(X)
pyplot.scatter(result[:, 0], result[:, 1])
words = list(model.wv.vocab)
for i, word in enumerate(words):
pyplot.annotate(word, xy=(result[i, 0], result[i, 1]))
pyplot.show()
plot_pca(model)
Node2Vec¶
Search strategies
- BFS: Breadth-first Sampling
- DFS: Depth-first Sampling
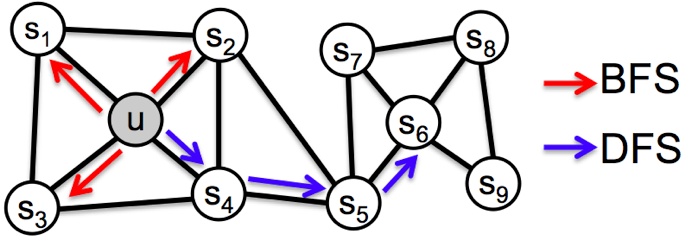
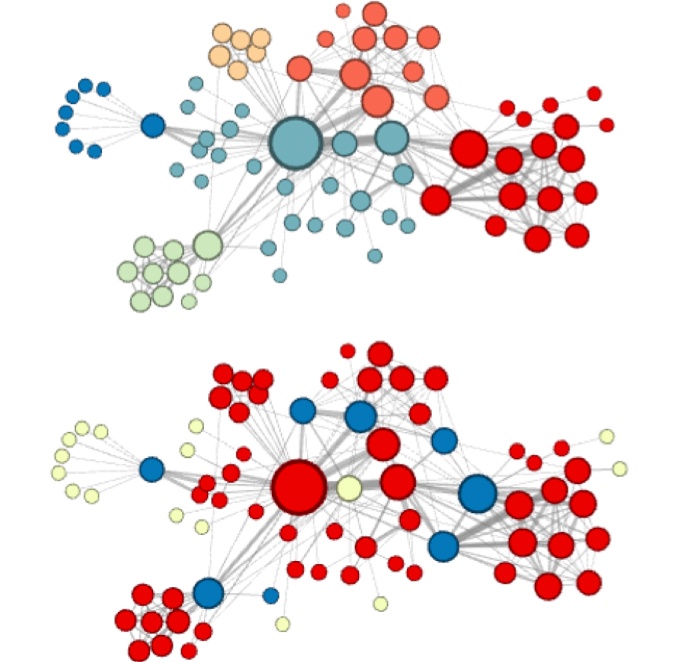
Node similarities in embedding space
- Homophily:
- Similarity in terms of the neighbouring of nodes
- Structural equivalence:
- Similarity in terms of structural roles; not emphasized on connectivity
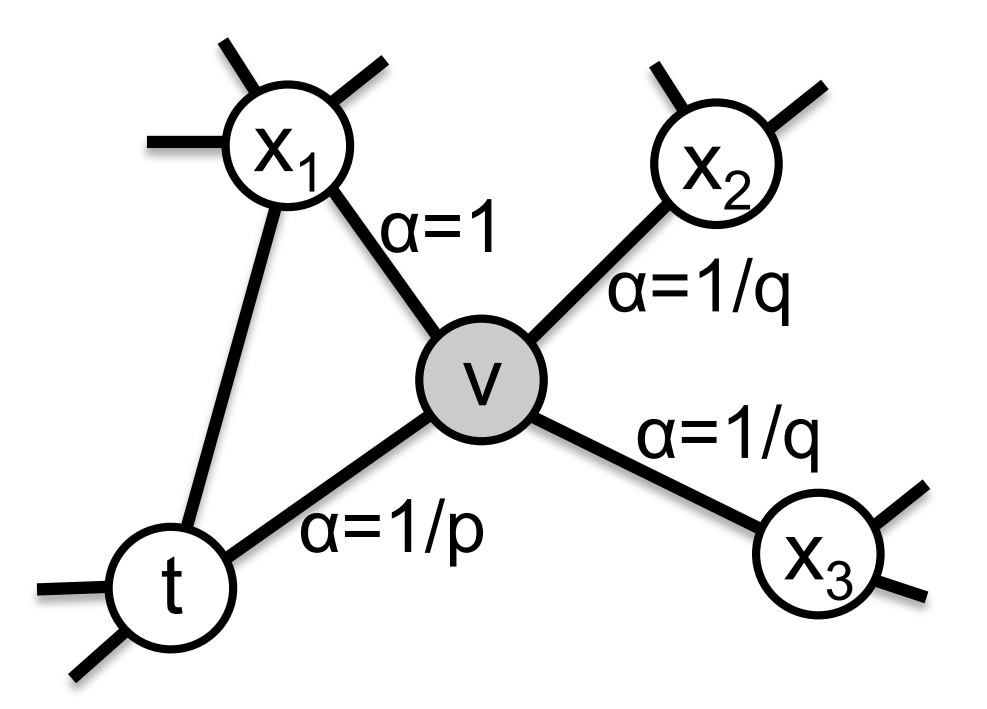
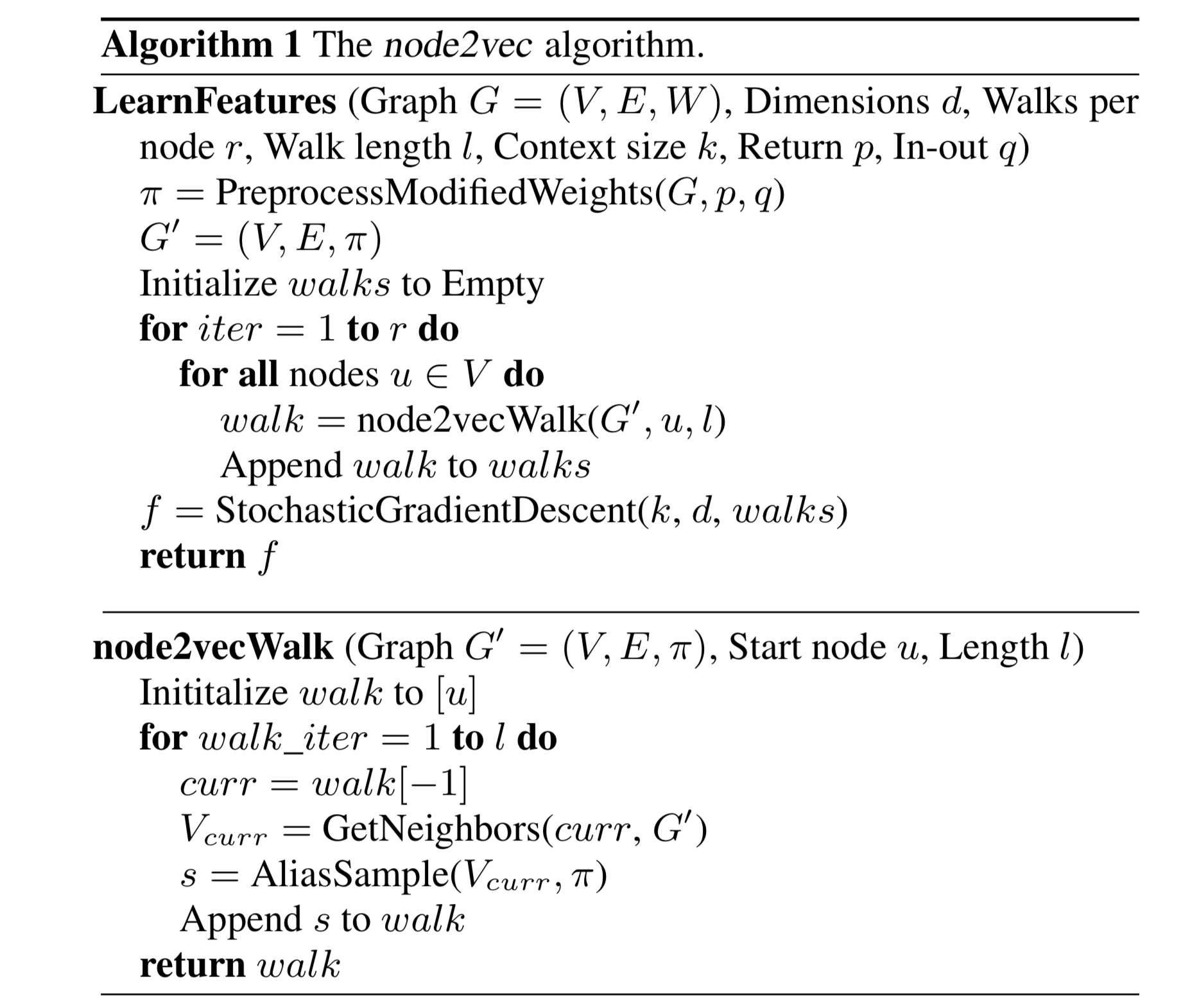
2nd order random walk:
- Mixture of BFS and DFS
- Search bias $\alpha$:
- $p$: likelihood of immediately revisiting a node
- $q$: larger $q$ biased towards reversion, smaller $q$ biased towards visiting new nodes
- semi-supervised learning: decide p, q from labeled nodes
In [13]:
from node2vec import Node2Vec
In [14]:
node2vec = Node2Vec(
nx_graph, dimensions=EMBEDDING_DIM, walk_length=WALK_LENGTH, num_walks=NUM_WALKS
)
Computing transition probabilities: 100%|██████████| 13/13 [00:00<00:00, 7282.75it/s] Generating walks (CPU: 1): 100%|██████████| 80/80 [00:01<00:00, 63.13it/s]
In [15]:
print(
"walks: \n", np.array(node2vec.walks),
"\n\n",
"shape: ", np.array(node2vec.walks).shape
)
walks: [['9' '10' '11' ... '3' '4' '5'] ['6' '3' '4' ... '2' '1' '2'] ['12' '11' '12' ... '1' '2' '6'] ... ['5' '4' '6' ... '8' '11' '8'] ['10' '11' '12' ... '3' '6' '3'] ['11' '8' '13' ... '8' '9' '10']] shape: (1040, 40)
In [16]:
model = node2vec.fit(window=WINDOW_SIZE, min_count=0)
In [17]:
model.wv.vocab
Out[17]:
{'9': <gensim.models.keyedvectors.Vocab at 0x1a1e4f6710>,
'10': <gensim.models.keyedvectors.Vocab at 0x1a1e4f6750>,
'11': <gensim.models.keyedvectors.Vocab at 0x1a1e4f6790>,
'8': <gensim.models.keyedvectors.Vocab at 0x1a1e4f6810>,
'7': <gensim.models.keyedvectors.Vocab at 0x1a1e4f6890>,
'6': <gensim.models.keyedvectors.Vocab at 0x1a1e4f6910>,
'2': <gensim.models.keyedvectors.Vocab at 0x1a1e4f6950>,
'5': <gensim.models.keyedvectors.Vocab at 0x1a1e4f6990>,
'4': <gensim.models.keyedvectors.Vocab at 0x1a1e4f6850>,
'3': <gensim.models.keyedvectors.Vocab at 0x1a1e4f68d0>,
'1': <gensim.models.keyedvectors.Vocab at 0x1a1e4f69d0>,
'12': <gensim.models.keyedvectors.Vocab at 0x1a1e4f6a10>,
'13': <gensim.models.keyedvectors.Vocab at 0x1a1e4f6a50>}
In [18]:
plot_pca(model)
EpiGraphDB¶
In [19]:
import json
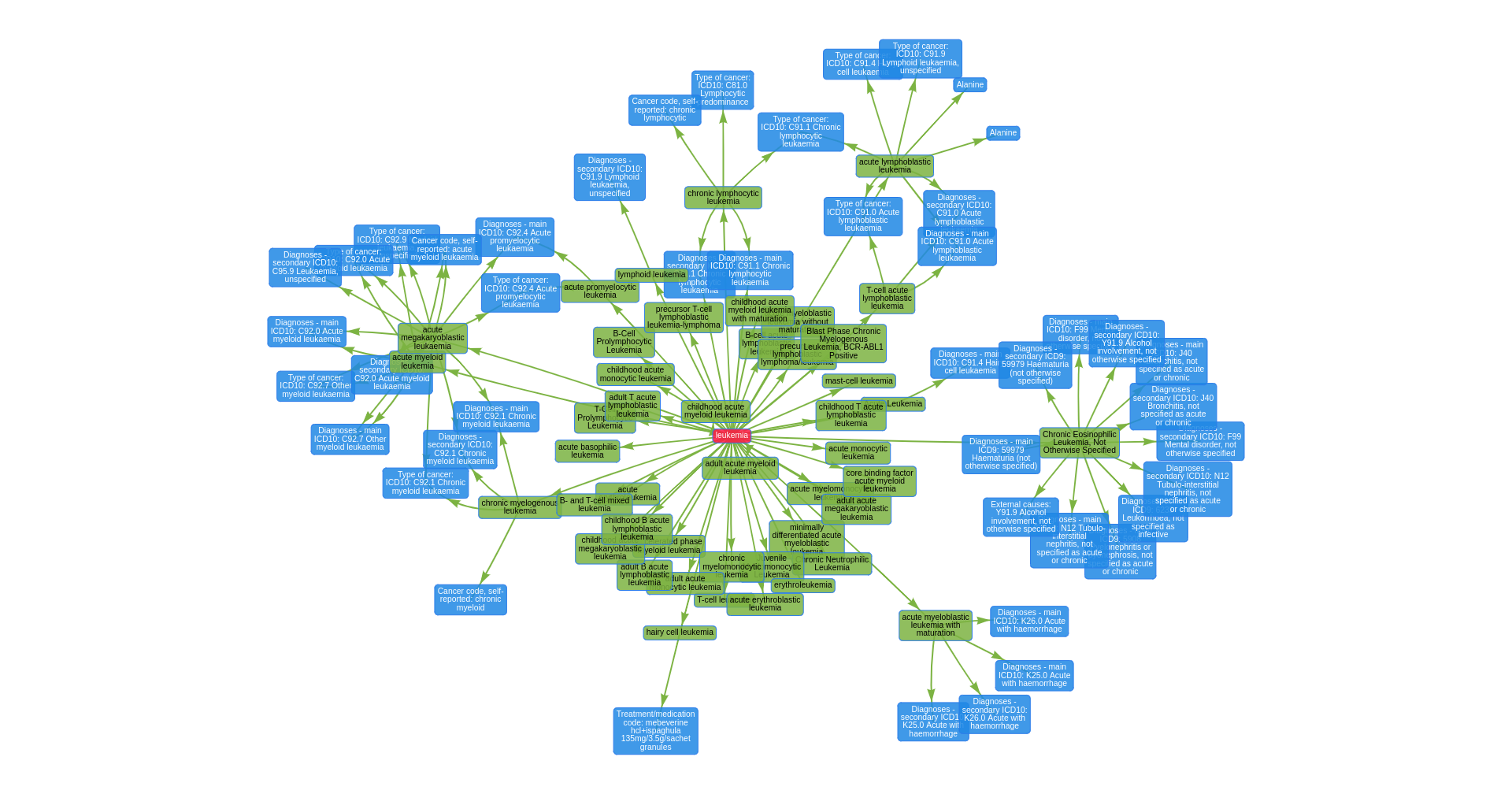
with open("assets/epigraphdb_efo_leukemia.json", "r") as f:
epigraphdb_data = json.load(f)
In [20]:
from pprint import pprint
pprint(epigraphdb_data["nodes"][:3])
pprint(epigraphdb_data["edges"][:3])
[{'id': 44,
'label': 'Diagnoses - secondary ICD10: Y91.9 Alcohol involvement, not '
'otherwise specified'},
{'id': 37,
'label': 'Diagnoses - secondary ICD9: 59979 Haematuria (not otherwise '
'specified)'},
{'id': 31,
'label': 'Diagnoses - main ICD10: J40 Bronchitis, not specified as acute or '
'chronic'}]
[{'from': 57, 'to': 69}, {'from': 57, 'to': 71}, {'from': 57, 'to': 70}]
In [21]:
epigraphdb_graph = nx.Graph()
epigraphdb_graph.add_nodes_from([item["id"] for item in epigraphdb_data["nodes"]])
epigraphdb_graph.add_edges_from([(item["from"], item["to"]) for item in epigraphdb_data["edges"]])
nx.draw(epigraphdb_graph, with_labels=True, node_color="white")
In [22]:
epigraphdb_node2vec = Node2Vec(
epigraphdb_graph, dimensions=EMBEDDING_DIM, walk_length=WALK_LENGTH, num_walks=NUM_WALKS
)
Computing transition probabilities: 100%|██████████| 91/91 [00:00<00:00, 7093.80it/s] Generating walks (CPU: 1): 100%|██████████| 80/80 [00:09<00:00, 8.67it/s]
In [23]:
epigraphdb_model = epigraphdb_node2vec.fit(window=WINDOW_SIZE, min_count=0)
In [24]:
plot_pca(epigraphdb_model)
In [25]:
epigraphdb_model.most_similar("24")
Out[25]:
[('45', 0.7994406819343567),
('2', 0.7983152866363525),
('20', 0.7964375019073486),
('47', 0.7630603909492493),
('8', 0.7575975656509399),
('13', 0.7530874013900757),
('50', 0.7519896626472473),
('1', 0.746828019618988),
('33', 0.7092547416687012),
('9', 0.7086656093597412)]
In [26]:
from sklearn.manifold import TSNE
def plot_tsne(model):
X = model[model.wv.vocab]
result = TSNE(n_components=2).fit_transform(X)
pyplot.scatter(result[:, 0], result[:, 1])
words = list(model.wv.vocab)
for i, word in enumerate(words):
pyplot.annotate(word, xy=(result[i, 0], result[i, 1]))
pyplot.show()
plot_tsne(epigraphdb_model)
References¶
- DeepWalk: https://arxiv.org/abs/1403.6652
- Node2Vec: https://cs.stanford.edu/~jure/pubs/node2vec-kdd16.pdf
- http://media.cs.tsinghua.edu.cn/~multimedia/cuipeng/papers/Network%20Representation-Tutorial.pdf
- https://medium.com/@_init_/an-illustrated-explanation-of-using-skipgram-to-encode-the-structure-of-a-graph-deepwalk-6220e304d71b
- https://towardsdatascience.com/graph-embeddings-the-summary-cc6075aba007
- https://towardsdatascience.com/node2vec-embeddings-for-graph-data-32a866340fef
- https://neo4j.com/online-summit/session/graph-embeddings-machine-learning-ml